Core Publications
ANTEC: Bioactive Scaffolds With Enhanced Supramolecular Motion Promote Recovery from Spinal Cord Injury
Alvarez, Z.; Edelbrock, A. N.; Sasselli, I. R.; Ortega, J. A.; Syrgiannis, Z.; Mirau, P. A.; Chen, F.; Qiu, R.; Chin, S.M.; Kiskinis, E.; Stupp, S. I.
Abstract
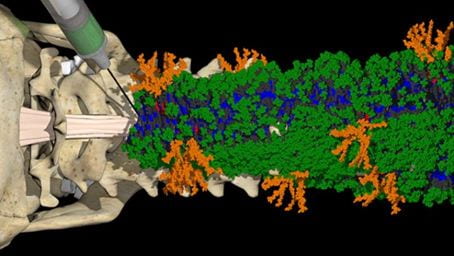 The signaling of cells by scaffolds of synthetic molecules that mimic proteins is known to be effective in the regeneration of tissues. Here, we describe peptide amphiphile supramolecular polymers containing two distinct signals and test them in a mouse model of severe spinal cord injury. One signal activates the transmembrane receptor β1-integrin and a second one activates the basic fibroblast growth factor 2 receptor. By mutating the peptide sequence of the amphiphilic monomers in nonbioactive domains, we intensified the motions of molecules within scaffold fibrils. This resulted in notable differences in vascular growth, axonal regeneration, myelination, survival of motor neurons, reduced gliosis, and functional recovery. We hypothesize that the signaling of cells by ensembles of molecules could be optimized by tuning their internal motions.
The signaling of cells by scaffolds of synthetic molecules that mimic proteins is known to be effective in the regeneration of tissues. Here, we describe peptide amphiphile supramolecular polymers containing two distinct signals and test them in a mouse model of severe spinal cord injury. One signal activates the transmembrane receptor β1-integrin and a second one activates the basic fibroblast growth factor 2 receptor. By mutating the peptide sequence of the amphiphilic monomers in nonbioactive domains, we intensified the motions of molecules within scaffold fibrils. This resulted in notable differences in vascular growth, axonal regeneration, myelination, survival of motor neurons, reduced gliosis, and functional recovery. We hypothesize that the signaling of cells by ensembles of molecules could be optimized by tuning their internal motions.
- The authors utilized ANTEC equipment including
- IncuCyte Live Cell Imager (obtained with OR/SQI funding, 2018)
- MCR 302 Rheometer (OR funding, 2016)
- Azure Chemiluminescent Gel Imager system (ReLODE, 2016)
- Cytation3 multimodal plate reader (OR funding, 2013)
- Lyophilizers
- Scientific Illustration service was provided by Mark Seniw
SPID: Micromachined Chip Scale Thermal Sensor for Thermal Imaging
 Gajendra S. Shekhawat, Srinivasan Ramachandran, Hossein Jiryaei Sharahi, Souravi Sarkar, Karl Hujsak, Yuan Li, Karl Hagglund, Seonghwan Kim, Gary Aden, Ami Chand, and Vinayak P. Dravid
Gajendra S. Shekhawat, Srinivasan Ramachandran, Hossein Jiryaei Sharahi, Souravi Sarkar, Karl Hujsak, Yuan Li, Karl Hagglund, Seonghwan Kim, Gary Aden, Ami Chand, and Vinayak P. Dravid
Abstract
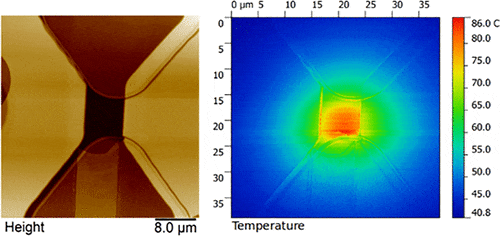
The lateral resolution of scanning thermal microscopy (SThM) has hitherto never approached that of mainstream atomic force microscopy, mainly due to poor performance of the thermal sensor. We have developed a thermocouple-based probe technology delivers excellent lateral resolution (∼20 nm), extended high-temperature measurements >700 °C without cantilever bending, and thermal sensitivity (∼0.04 °C). The origin of significantly improved figures-of-merit lies in the probe design that consists of a hollow silicon tip integrated with a vertically oriented thermocouple sensor at the apex (low thermal mass) which interacts with the sample through a metallic nanowire (50 nm diameter), thereby achieving high lateral resolution. The nanoscale pitch and extremely small thermal mass of the probe promise significant improvements over existing methods and wide range of applications in several fields including semiconductor industry, biomedical imaging, and data storage.
CAM: Reflectance Imaging for Visualization of Unlabeled Structures Using Nikon A1 and N-SIM
 Dr. Emily J Guggenheim and Dr. Joshua Z Rappoport
Dr. Emily J Guggenheim and Dr. Joshua Z Rappoport
Abstract
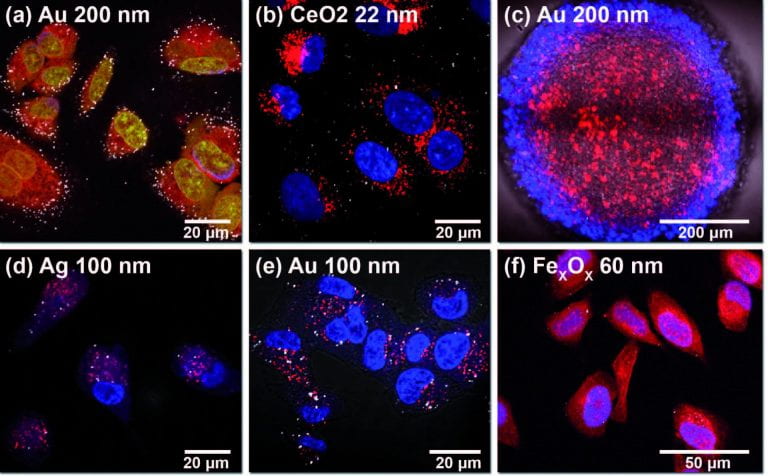
Reflectance imaging allows the user to form an intensity image from light backscattered by the sample. Highly reflective markers, including a variety of nanoparticles, allows for imaging with very high signal-to-noise and virtually free of photobleaching, ideal for both confocal and structured illumination microscopies.
IMSERC: Best Practices for the Synthesis, Activation, and Characterization of Metal-Organic Frameworks
 Ashlee J. Howarth, Aaron W. Peters, Nicolaas A. Vermeulen, Timothy C. Wang, Joseph T. Hupp, and Omar K. Farha
Ashlee J. Howarth, Aaron W. Peters, Nicolaas A. Vermeulen, Timothy C. Wang, Joseph T. Hupp, and Omar K. Farha
Abstract
Data collected in the Integrated Molecular Structure Education and Research Center (IMSERC) was featured on the cover of Chemistry of Materials describing best practices for metal-organic framework materials (MOF’s). MOF’s have record breaking surface areas and applications in gas storage and catalysis applications. IMSERC single crystal X-Ray diffraction, powder X-Ray diffraction, mass spectrometry and NMR spectrometry were used to characterize bulk materials and novel organic linkers. The ability to collect single crystal XRD data on crystals with <100mm/side has rapidly increased the rate of development in this field.
NUSeq: Next-generation sequencing data analysis
 Xinkun Wang
Xinkun Wang
Abstract
The development of massively parallel genome sequencing technologies has led to a paradigm shift in biomedical research, and the emergence of precision medicine. Scientists and clinicians are now able to see how human diseases and phenotypic changes are connected to DNA sequence changes, genome structural changes, and epigenomic abnormality. Next-Generation Sequencing Data Analysis, authored by the director of the Northwestern University NUSeq Core Facility, provides a practical guide to applying these powerful genome technologies to major areas of biomedical research.
The book walks readers through multiple stages of NGS data generation and analysis in an easy-to-follow fashion. It covers every step in each stage, from the planning stage of experimental design, sample processing, sequencing strategy formulation, the early stage of base calling, reads quality check and data preprocessing to the intermediate stage of mapping reads to a reference genome and normalization to more advanced stages specific to each application. All major applications of NGS are covered, including: RNA-seq: gene expression analysis, Genome re-sequencing: genotyping and variant discovery, De novo genome assembly, ChIP-seq: study of DNA–protein interaction, DNA methyl-seq: epigenomic regulation, Microbiome sequencing.
HTAL: Discovery of novel Mnk inhibitors using mutation-based induced-fit virtual high-throughput screening
 Mishra RK, Clutter MR, Blyth GT, Kosciuczuk EM, Blackburn AZ, Beauchamp EM, Schiltz GE, Platanias LC.
Mishra RK, Clutter MR, Blyth GT, Kosciuczuk EM, Blackburn AZ, Beauchamp EM, Schiltz GE, Platanias LC.
Abstract
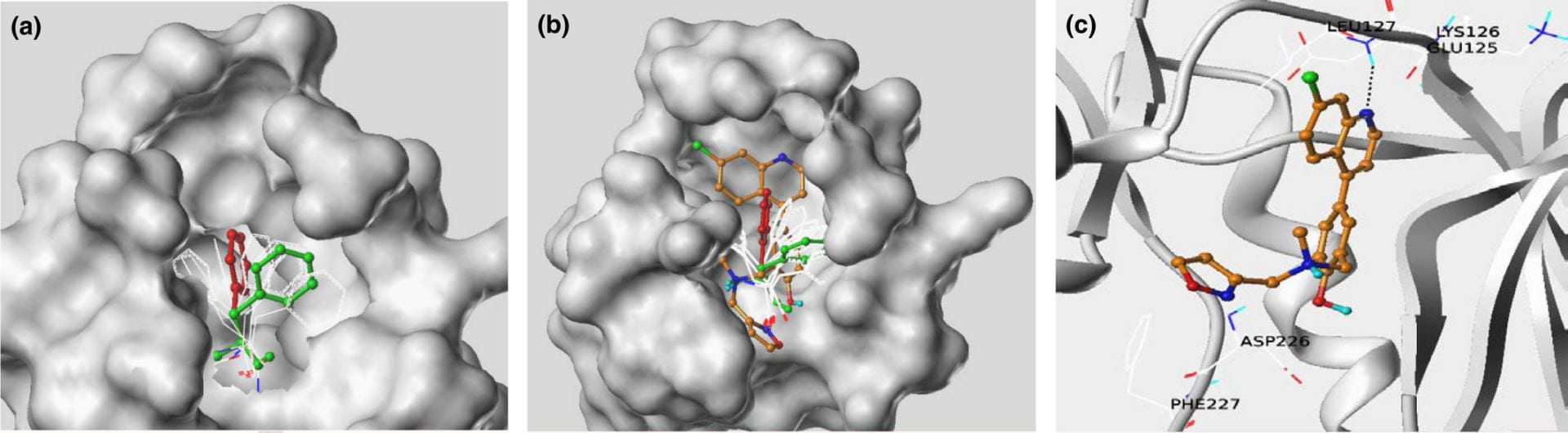
Mnk kinases (Mnk1 and 2) are downstream effectors of Map kinase pathways and regulate phosphorylation of eukaryotic initiation factor 4E (eIF4E). Engagement of the Mnk pathway is critical in acute myeloid leukemia (AML) leukemogenesis and Mnk inhibitors have potent antileukemic properties in vitro and in vivo, suggesting that targeting Mnk kinases may provide a novel approach for treating AML. Here, we report the development and application of a mutation-based induced-fit (M-IFD) in silico screen to identify novel Mnk inhibitors. The Mnk1 structure was modeled by temporarily mutating an amino acid that obstructs the ATP-binding site in the Mnk1 crystal structure while carrying out docking simulations of known inhibitors. The hit compounds display activity in Mnk biochemical and cellular assays, including acute myeloid leukemia progenitors. This approach will enable further rational structure-based drug design of new Mnk inhibitors and potentially novel ways of therapeutically targeting this kinase.